做兼职的网站有哪些工作内容成都有实力的seo团队
基于R 4.2.2版本演示
一、写在前面
有不少大佬问做机器学习分类能不能用R语言,不想学Python咯。
答曰:可!用GPT或者Kimi转一下就得了呗。
加上最近也没啥内容写了,就帮各位搬运一下吧。
二、R代码实现决策树分类
(1)导入数据
我习惯用RStudio自带的导入功能:
(2)建立决策树模型(默认参数)
# Load necessary libraries
library(caret)
library(pROC)
library(ggplot2)# Assume 'data' is your dataframe containing the data
# Set seed to ensure reproducibility
set.seed(123)# Split data into training and validation sets (80% training, 20% validation)
trainIndex <- createDataPartition(data$X, p = 0.8, list = FALSE)
trainData <- data[trainIndex, ]
validData <- data[-trainIndex, ]# Convert the target variable to a factor for classification
trainData$X <- as.factor(trainData$X)
validData$X <- as.factor(validData$X)# Define control method for training with cross-validation
trainControl <- trainControl(method = "cv", number = 10)
# Fit Decision Tree model on the training set
model <- train(X ~ ., data = trainData, method = "rpart", trControl = trainControl)# Print the best parameters found by the model
best_params <- model$bestTune
cat("The best parameters found are:\n")
print(best_params)# Predict on the training and validation sets
trainPredict <- predict(model, trainData, type = "prob")[,2]
validPredict <- predict(model, validData, type = "prob")[,2]# Convert true values to factor for ROC analysis
trainData$X <- as.factor(trainData$X)
validData$X <- as.factor(validData$X)# Calculate ROC curves and AUC values
trainRoc <- roc(response = trainData$X, predictor = trainPredict)
validRoc <- roc(response = validData$X, predictor = validPredict)# Plot ROC curves with AUC values
ggplot(data = data.frame(fpr = trainRoc$specificities, tpr = trainRoc$sensitivities), aes(x = 1 - fpr, y = tpr)) +geom_line(color = "blue") +geom_area(alpha = 0.2, fill = "blue") +geom_abline(slope = 1, intercept = 0, linetype = "dashed", color = "black") +ggtitle("Training ROC Curve") +xlab("False Positive Rate") +ylab("True Positive Rate") +annotate("text", x = 0.5, y = 0.1, label = paste("Training AUC =", round(auc(trainRoc), 2)), hjust = 0.5, color = "blue")ggplot(data = data.frame(fpr = validRoc$specificities, tpr = validRoc$sensitivities), aes(x = 1 - fpr, y = tpr)) +geom_line(color = "red") +geom_area(alpha = 0.2, fill = "red") +geom_abline(slope = 1, intercept = 0, linetype = "dashed", color = "black") +ggtitle("Validation ROC Curve") +xlab("False Positive Rate") +ylab("True Positive Rate") +annotate("text", x = 0.5, y = 0.2, label = paste("Validation AUC =", round(auc(validRoc), 2)), hjust = 0.5, color = "red")# Calculate confusion matrices based on 0.5 cutoff for probability
confMatTrain <- table(trainData$X, trainPredict >= 0.5)
confMatValid <- table(validData$X, validPredict >= 0.5)# Function to plot confusion matrix using ggplot2
plot_confusion_matrix <- function(conf_mat, dataset_name) {conf_mat_df <- as.data.frame(as.table(conf_mat))colnames(conf_mat_df) <- c("Actual", "Predicted", "Freq")p <- ggplot(data = conf_mat_df, aes(x = Predicted, y = Actual, fill = Freq)) +geom_tile(color = "white") +geom_text(aes(label = Freq), vjust = 1.5, color = "black", size = 5) +scale_fill_gradient(low = "white", high = "steelblue") +labs(title = paste("Confusion Matrix -", dataset_name, "Set"), x = "Predicted Class", y = "Actual Class") +theme_minimal() +theme(axis.text.x = element_text(angle = 45, hjust = 1), plot.title = element_text(hjust = 0.5))print(p)
}
# Now call the function to plot and display the confusion matrices
plot_confusion_matrix(confMatTrain, "Training")
plot_confusion_matrix(confMatValid, "Validation")# Extract values for calculations
a_train <- confMatTrain[1, 1]
b_train <- confMatTrain[1, 2]
c_train <- confMatTrain[2, 1]
d_train <- confMatTrain[2, 2]a_valid <- confMatValid[1, 1]
b_valid <- confMatValid[1, 2]
c_valid <- confMatValid[2, 1]
d_valid <- confMatValid[2, 2]# Training Set Metrics
acc_train <- (a_train + d_train) / sum(confMatTrain)
error_rate_train <- 1 - acc_train
sen_train <- d_train / (d_train + c_train)
sep_train <- a_train / (a_train + b_train)
precision_train <- d_train / (b_train + d_train)
F1_train <- (2 * precision_train * sen_train) / (precision_train + sen_train)
MCC_train <- (d_train * a_train - b_train * c_train) / sqrt((d_train + b_train) * (d_train + c_train) * (a_train + b_train) * (a_train + c_train))
auc_train <- roc(response = trainData$X, predictor = trainPredict)$auc# Validation Set Metrics
acc_valid <- (a_valid + d_valid) / sum(confMatValid)
error_rate_valid <- 1 - acc_valid
sen_valid <- d_valid / (d_valid + c_valid)
sep_valid <- a_valid / (a_valid + b_valid)
precision_valid <- d_valid / (b_valid + d_valid)
F1_valid <- (2 * precision_valid * sen_valid) / (precision_valid + sen_valid)
MCC_valid <- (d_valid * a_valid - b_valid * c_valid) / sqrt((d_valid + b_valid) * (d_valid + c_valid) * (a_valid + b_valid) * (a_valid + c_valid))
auc_valid <- roc(response = validData$X, predictor = validPredict)$auc# Print Metrics
cat("Training Metrics\n")
cat("Accuracy:", acc_train, "\n")
cat("Error Rate:", error_rate_train, "\n")
cat("Sensitivity:", sen_train, "\n")
cat("Specificity:", sep_train, "\n")
cat("Precision:", precision_train, "\n")
cat("F1 Score:", F1_train, "\n")
cat("MCC:", MCC_train, "\n")
cat("AUC:", auc_train, "\n\n")cat("Validation Metrics\n")
cat("Accuracy:", acc_valid, "\n")
cat("Error Rate:", error_rate_valid, "\n")
cat("Sensitivity:", sen_valid, "\n")
cat("Specificity:", sep_valid, "\n")
cat("Precision:", precision_valid, "\n")
cat("F1 Score:", F1_valid, "\n")
cat("MCC:", MCC_valid, "\n")
cat("AUC:", auc_valid, "\n")在R语言中,还是使用caret包来训练决策树模型,可以调整多种参数来优化模型的性能:
①cp (Complexity Parameter): 用来控制树的生长。cp 值越大,生成的模型越简单。如果 cp 设置得太高,可能导致模型欠拟合。
②maxdepth: 决定了树的最大深度。较深的树可以更好地捕捉数据中的复杂关系,但也可能导致过拟合。
③minsplit: 定义了节点在尝试分裂之前所需的最小样本数。增加这个值可以让树更加稳健,但也可能导致欠拟合。
④minbucket: 叶节点最少包含的样本数。这个参数可以帮助防止模型过于复杂,从而避免过拟合。
结果输出(默认参数):
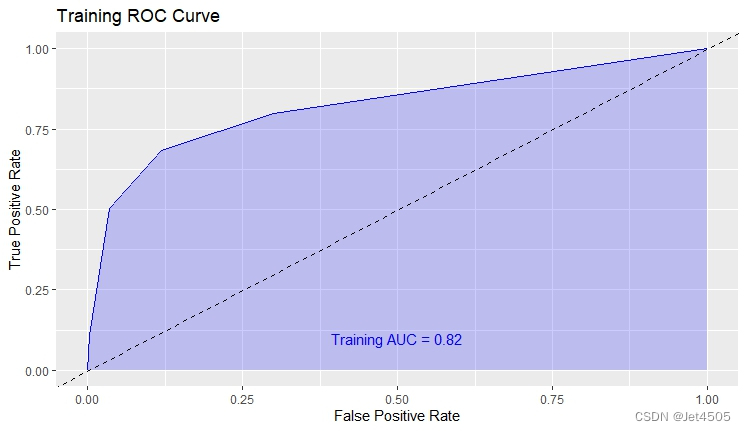
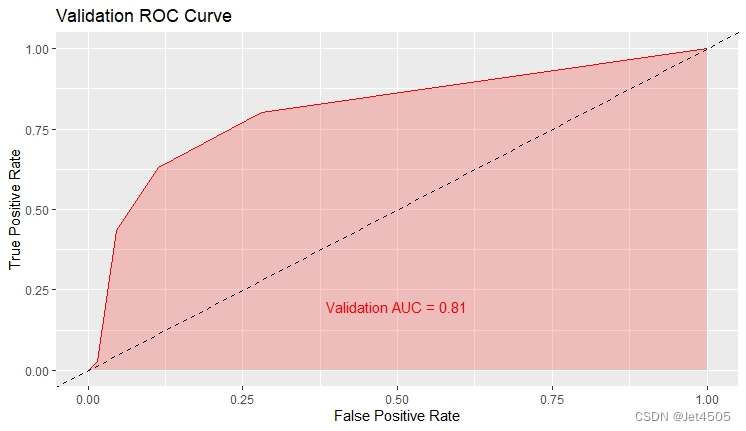
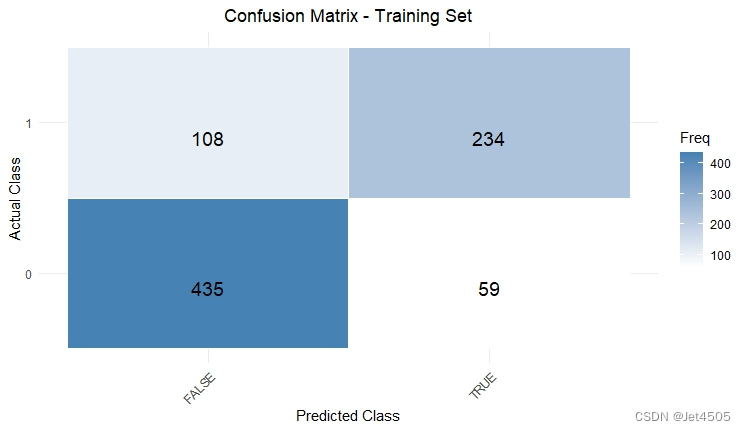
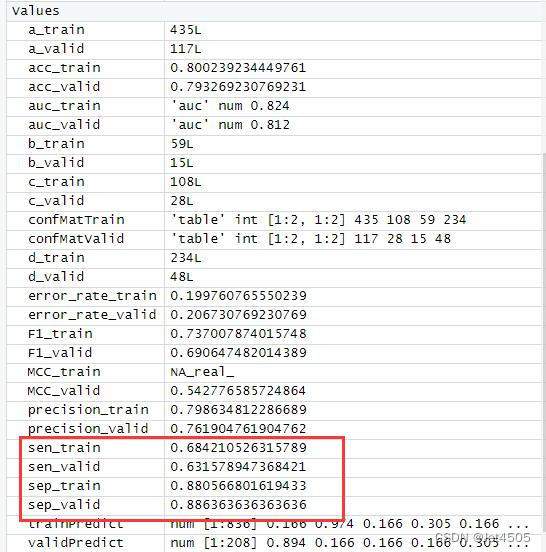
在默认参数中,caret包只会默默帮我们找几个合适的cp值进行测试。其他三个参数就一个默认值。
三、决策树调参方法(仅cp值)
如前所述,决策树的关键参数就是有4个,但是caret包做网格搜索的话,只能提供对于cp值的遍历,其余三个不提供。
比如我设置cp值从0.001到0.1,步长是0.001;而maxdepth = 20, minsplit = 10, minbucket = 10:
# Load necessary libraries
library(caret)
library(pROC)
library(ggplot2)
library(rpart)# Assume 'data' is your dataframe containing the data
# Set seed to ensure reproducibility
set.seed(123)# Split data into training and validation sets (80% training, 20% validation)
trainIndex <- createDataPartition(data$X, p = 0.8, list = FALSE)
trainData <- data[trainIndex, ]
validData <- data[-trainIndex, ]# Convert the target variable to a factor for classification
trainData$X <- as.factor(trainData$X)
validData$X <- as.factor(validData$X)# 定义交叉验证控制方法
trainControl <- trainControl(method = "cv", number = 10)# 定义参数网格,只包括 cp
tuneGrid <- expand.grid(cp = seq(0.001, 0.1, by = 0.001))# 定义 rpart.control,固定其他参数
rpartControl <- rpart.control(maxdepth = 20, minsplit = 10, minbucket = 10)# 使用 rpart 方法训练决策树模型
model <- train(X ~ ., data = trainData, method = "rpart", trControl = trainControl, tuneGrid = tuneGrid,control = rpartControl)# 打印找到的最佳参数
best_params <- model$bestTune
cat("The best parameters found are:\n")
print(best_params)# Predict on the training and validation sets
trainPredict <- predict(model, trainData, type = "prob")[,2]
validPredict <- predict(model, validData, type = "prob")[,2]# Convert true values to factor for ROC analysis
trainData$X <- as.factor(trainData$X)
validData$X <- as.factor(validData$X)# Calculate ROC curves and AUC values
trainRoc <- roc(response = trainData$X, predictor = trainPredict)
validRoc <- roc(response = validData$X, predictor = validPredict)# Plot ROC curves with AUC values
ggplot(data = data.frame(fpr = trainRoc$specificities, tpr = trainRoc$sensitivities), aes(x = 1 - fpr, y = tpr)) +geom_line(color = "blue") +geom_area(alpha = 0.2, fill = "blue") +geom_abline(slope = 1, intercept = 0, linetype = "dashed", color = "black") +ggtitle("Training ROC Curve") +xlab("False Positive Rate") +ylab("True Positive Rate") +annotate("text", x = 0.5, y = 0.1, label = paste("Training AUC =", round(auc(trainRoc), 2)), hjust = 0.5, color = "blue")ggplot(data = data.frame(fpr = validRoc$specificities, tpr = validRoc$sensitivities), aes(x = 1 - fpr, y = tpr)) +geom_line(color = "red") +geom_area(alpha = 0.2, fill = "red") +geom_abline(slope = 1, intercept = 0, linetype = "dashed", color = "black") +ggtitle("Validation ROC Curve") +xlab("False Positive Rate") +ylab("True Positive Rate") +annotate("text", x = 0.5, y = 0.2, label = paste("Validation AUC =", round(auc(validRoc), 2)), hjust = 0.5, color = "red")# Calculate confusion matrices based on 0.5 cutoff for probability
confMatTrain <- table(trainData$X, trainPredict >= 0.5)
confMatValid <- table(validData$X, validPredict >= 0.5)# Function to plot confusion matrix using ggplot2
plot_confusion_matrix <- function(conf_mat, dataset_name) {conf_mat_df <- as.data.frame(as.table(conf_mat))colnames(conf_mat_df) <- c("Actual", "Predicted", "Freq")p <- ggplot(data = conf_mat_df, aes(x = Predicted, y = Actual, fill = Freq)) +geom_tile(color = "white") +geom_text(aes(label = Freq), vjust = 1.5, color = "black", size = 5) +scale_fill_gradient(low = "white", high = "steelblue") +labs(title = paste("Confusion Matrix -", dataset_name, "Set"), x = "Predicted Class", y = "Actual Class") +theme_minimal() +theme(axis.text.x = element_text(angle = 45, hjust = 1), plot.title = element_text(hjust = 0.5))print(p)
}
# Now call the function to plot and display the confusion matrices
plot_confusion_matrix(confMatTrain, "Training")
plot_confusion_matrix(confMatValid, "Validation")# Extract values for calculations
a_train <- confMatTrain[1, 1]
b_train <- confMatTrain[1, 2]
c_train <- confMatTrain[2, 1]
d_train <- confMatTrain[2, 2]a_valid <- confMatValid[1, 1]
b_valid <- confMatValid[1, 2]
c_valid <- confMatValid[2, 1]
d_valid <- confMatValid[2, 2]# Training Set Metrics
acc_train <- (a_train + d_train) / sum(confMatTrain)
error_rate_train <- 1 - acc_train
sen_train <- d_train / (d_train + c_train)
sep_train <- a_train / (a_train + b_train)
precision_train <- d_train / (b_train + d_train)
F1_train <- (2 * precision_train * sen_train) / (precision_train + sen_train)
MCC_train <- (d_train * a_train - b_train * c_train) / sqrt((d_train + b_train) * (d_train + c_train) * (a_train + b_train) * (a_train + c_train))
auc_train <- roc(response = trainData$X, predictor = trainPredict)$auc# Validation Set Metrics
acc_valid <- (a_valid + d_valid) / sum(confMatValid)
error_rate_valid <- 1 - acc_valid
sen_valid <- d_valid / (d_valid + c_valid)
sep_valid <- a_valid / (a_valid + b_valid)
precision_valid <- d_valid / (b_valid + d_valid)
F1_valid <- (2 * precision_valid * sen_valid) / (precision_valid + sen_valid)
MCC_valid <- (d_valid * a_valid - b_valid * c_valid) / sqrt((d_valid + b_valid) * (d_valid + c_valid) * (a_valid + b_valid) * (a_valid + c_valid))
auc_valid <- roc(response = validData$X, predictor = validPredict)$auc# Print Metrics
cat("Training Metrics\n")
cat("Accuracy:", acc_train, "\n")
cat("Error Rate:", error_rate_train, "\n")
cat("Sensitivity:", sen_train, "\n")
cat("Specificity:", sep_train, "\n")
cat("Precision:", precision_train, "\n")
cat("F1 Score:", F1_train, "\n")
cat("MCC:", MCC_train, "\n")
cat("AUC:", auc_train, "\n\n")cat("Validation Metrics\n")
cat("Accuracy:", acc_valid, "\n")
cat("Error Rate:", error_rate_valid, "\n")
cat("Sensitivity:", sen_valid, "\n")
cat("Specificity:", sep_valid, "\n")
cat("Precision:", precision_valid, "\n")
cat("F1 Score:", F1_valid, "\n")
cat("MCC:", MCC_valid, "\n")
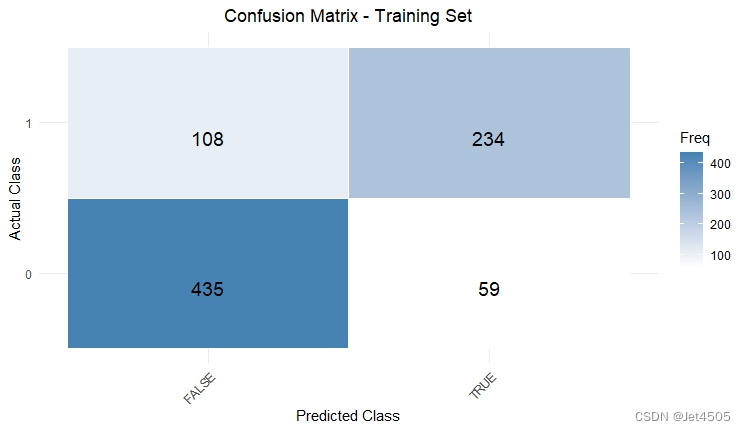
cat("AUC:", auc_valid, "\n")结果输出:
似乎好了一点了,那么,如果我先把其余三个参数也纳入遍历呢?那只能用循环语句了。
四、决策树调参方法(4个值)
设置cp值从0.001到0.1,步长是0.001;maxdepth从10到30,步长是5;minsplit取值10、20、30、40;minbucket取值5、10、15、20:
# Load necessary libraries
library(caret)
library(pROC)
library(ggplot2)
library(rpart)# Assume 'data' is your dataframe containing the data
# Set seed to ensure reproducibility
set.seed(123)# Split data into training and validation sets (80% training, 20% validation)
trainIndex <- createDataPartition(data$X, p = 0.8, list = FALSE)
trainData <- data[trainIndex, ]
validData <- data[-trainIndex, ]# Convert the target variable to a factor for classification
trainData$X <- as.factor(trainData$X)
validData$X <- as.factor(validData$X)# 参数网格定义
cp_values <- seq(0.001, 0.01, by = 0.001)
maxdepth_values <- seq(10, 30, by = 5)
minsplit_values <- c(10, 20, 30, 40)
minbucket_values <- c(5, 10, 15, 20)# 用于存储结果的列表
results <- list()# 网格搜索实现
for (cp in cp_values) {for (maxdepth in maxdepth_values) {for (minsplit in minsplit_values) {for (minbucket in minbucket_values) {# 训练模型model <- rpart(X ~ ., data = trainData, control = rpart.control(cp = cp, maxdepth = maxdepth,minsplit = minsplit, minbucket = minbucket))# 预测验证集predictions <- predict(model, validData, type = "class")# 计算性能指标,这里使用准确度accuracy <- sum(predictions == validData$X) / nrow(validData)# 存储结果results[[length(results) + 1]] <- list(cp = cp, maxdepth = maxdepth, minsplit = minsplit, minbucket = minbucket,accuracy = accuracy)}}}
}# 找到最高准确度的模型参数
best_model <- results[[which.max(sapply(results, function(x) x$accuracy))]]
print(best_model)# Predict on the training and validation sets
trainPredict <- predict(model, trainData, type = "prob")[,2]
validPredict <- predict(model, validData, type = "prob")[,2]# Convert true values to factor for ROC analysis
trainData$X <- as.factor(trainData$X)
validData$X <- as.factor(validData$X)# Calculate ROC curves and AUC values
trainRoc <- roc(response = trainData$X, predictor = trainPredict)
validRoc <- roc(response = validData$X, predictor = validPredict)# Plot ROC curves with AUC values
ggplot(data = data.frame(fpr = trainRoc$specificities, tpr = trainRoc$sensitivities), aes(x = 1 - fpr, y = tpr)) +geom_line(color = "blue") +geom_area(alpha = 0.2, fill = "blue") +geom_abline(slope = 1, intercept = 0, linetype = "dashed", color = "black") +ggtitle("Training ROC Curve") +xlab("False Positive Rate") +ylab("True Positive Rate") +annotate("text", x = 0.5, y = 0.1, label = paste("Training AUC =", round(auc(trainRoc), 2)), hjust = 0.5, color = "blue")ggplot(data = data.frame(fpr = validRoc$specificities, tpr = validRoc$sensitivities), aes(x = 1 - fpr, y = tpr)) +geom_line(color = "red") +geom_area(alpha = 0.2, fill = "red") +geom_abline(slope = 1, intercept = 0, linetype = "dashed", color = "black") +ggtitle("Validation ROC Curve") +xlab("False Positive Rate") +ylab("True Positive Rate") +annotate("text", x = 0.5, y = 0.2, label = paste("Validation AUC =", round(auc(validRoc), 2)), hjust = 0.5, color = "red")# Calculate confusion matrices based on 0.5 cutoff for probability
confMatTrain <- table(trainData$X, trainPredict >= 0.5)
confMatValid <- table(validData$X, validPredict >= 0.5)# Function to plot confusion matrix using ggplot2
plot_confusion_matrix <- function(conf_mat, dataset_name) {conf_mat_df <- as.data.frame(as.table(conf_mat))colnames(conf_mat_df) <- c("Actual", "Predicted", "Freq")p <- ggplot(data = conf_mat_df, aes(x = Predicted, y = Actual, fill = Freq)) +geom_tile(color = "white") +geom_text(aes(label = Freq), vjust = 1.5, color = "black", size = 5) +scale_fill_gradient(low = "white", high = "steelblue") +labs(title = paste("Confusion Matrix -", dataset_name, "Set"), x = "Predicted Class", y = "Actual Class") +theme_minimal() +theme(axis.text.x = element_text(angle = 45, hjust = 1), plot.title = element_text(hjust = 0.5))print(p)
}
# Now call the function to plot and display the confusion matrices
plot_confusion_matrix(confMatTrain, "Training")
plot_confusion_matrix(confMatValid, "Validation")# Extract values for calculations
a_train <- confMatTrain[1, 1]
b_train <- confMatTrain[1, 2]
c_train <- confMatTrain[2, 1]
d_train <- confMatTrain[2, 2]a_valid <- confMatValid[1, 1]
b_valid <- confMatValid[1, 2]
c_valid <- confMatValid[2, 1]
d_valid <- confMatValid[2, 2]# Training Set Metrics
acc_train <- (a_train + d_train) / sum(confMatTrain)
error_rate_train <- 1 - acc_train
sen_train <- d_train / (d_train + c_train)
sep_train <- a_train / (a_train + b_train)
precision_train <- d_train / (b_train + d_train)
F1_train <- (2 * precision_train * sen_train) / (precision_train + sen_train)
MCC_train <- (d_train * a_train - b_train * c_train) / sqrt((d_train + b_train) * (d_train + c_train) * (a_train + b_train) * (a_train + c_train))
auc_train <- roc(response = trainData$X, predictor = trainPredict)$auc# Validation Set Metrics
acc_valid <- (a_valid + d_valid) / sum(confMatValid)
error_rate_valid <- 1 - acc_valid
sen_valid <- d_valid / (d_valid + c_valid)
sep_valid <- a_valid / (a_valid + b_valid)
precision_valid <- d_valid / (b_valid + d_valid)
F1_valid <- (2 * precision_valid * sen_valid) / (precision_valid + sen_valid)
MCC_valid <- (d_valid * a_valid - b_valid * c_valid) / sqrt((d_valid + b_valid) * (d_valid + c_valid) * (a_valid + b_valid) * (a_valid + c_valid))
auc_valid <- roc(response = validData$X, predictor = validPredict)$auc# Print Metrics
cat("Training Metrics\n")
cat("Accuracy:", acc_train, "\n")
cat("Error Rate:", error_rate_train, "\n")
cat("Sensitivity:", sen_train, "\n")
cat("Specificity:", sep_train, "\n")
cat("Precision:", precision_train, "\n")
cat("F1 Score:", F1_train, "\n")
cat("MCC:", MCC_train, "\n")
cat("AUC:", auc_train, "\n\n")cat("Validation Metrics\n")
cat("Accuracy:", acc_valid, "\n")
cat("Error Rate:", error_rate_valid, "\n")
cat("Sensitivity:", sen_valid, "\n")
cat("Specificity:", sep_valid, "\n")
cat("Precision:", precision_valid, "\n")
cat("F1 Score:", F1_valid, "\n")
cat("MCC:", MCC_valid, "\n")
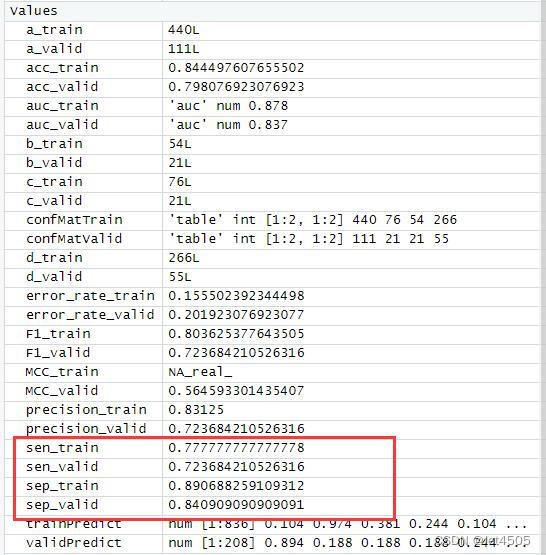
cat("AUC:", auc_valid, "\n")结果输出:
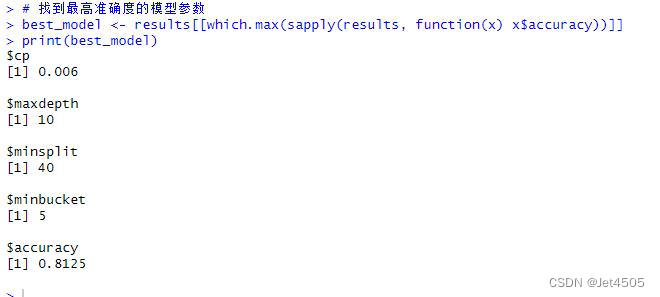
以上是找到的相对最优参数组合,看看具体性能:
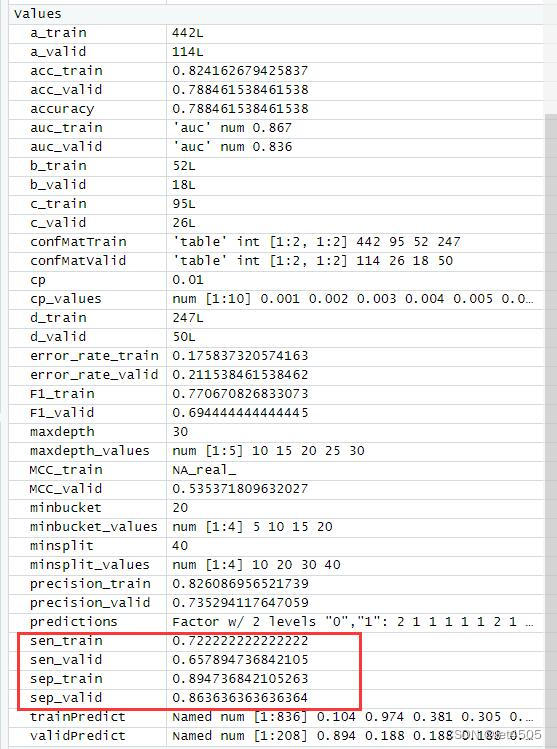
哈哈,又给调回去了,矫枉过正。思路就是这么个思路,大家自行食用了。
五、最后
看到这里,我觉得还是Python的sk-learn提供的调参简单些,至少不用写循环。
数据嘛:
链接:https://pan.baidu.com/s/1rEf6JZyzA1ia5exoq5OF7g?pwd=x8xm
提取码:x8xm
